Project members falling into the FGC5494 subclades can improve the data on this and the Pedigrees page through EXPERT LEVEL PARTICIPATION. Project cluster numbers for these members starting about 2800.
R1b subclade DF13 is the parent of several major haplogroups with significant representation in the British Isles. Among those major haplogroups under DF13 are some of what were called the "Gaelic" ones - Dal Cais, North Irish, South Irish, etc. Those groups, which turned up in early yDNA research, are now better known by their correlative SNPs: R-L226, R-M222, R-CTS4466, etc. At one time it was thought those groups in particular originated within Ireland. That view has shifted somewhat.
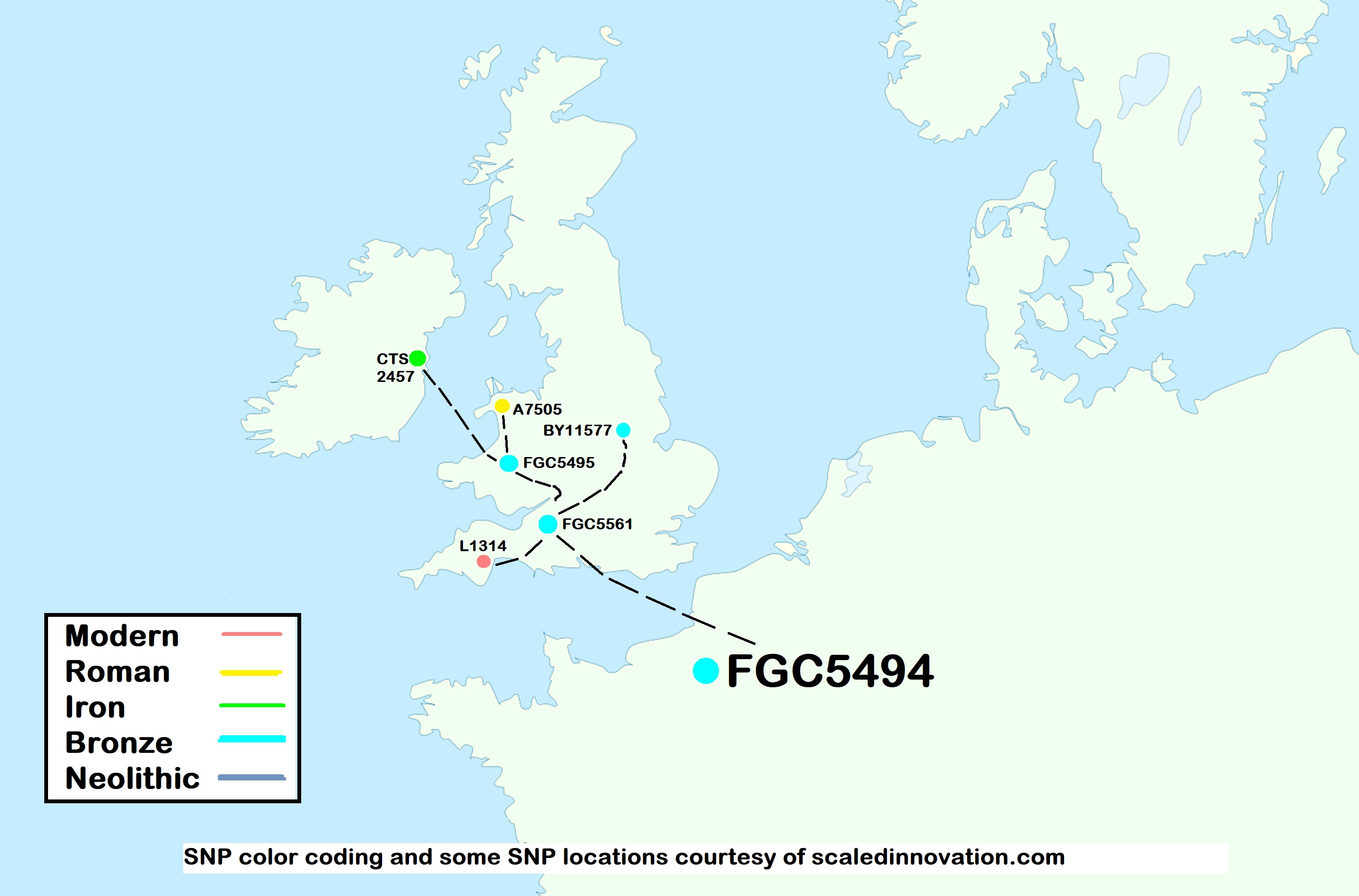
FGC5494 is is well dispersed in England, Scotland, Wales, Isle of Man, and Ireland, with a smattering on the European continent. FGC5494 probably entered the British Isles from the continent. Some have claimed Northern Frankish origins. Project members willing to commit financially to more extensive Y testing of STRs and SNPs should join the FGC5494 Haplogroup project.
Current estimates show a FGC5494 TMRCA date of roughly about 2500 BCE. The formation date of the FGC5494 mutation is about the same. FGC5494 has been found in ancient sites on the continent.
SNP Calendar / Timeline 
CLICK the further analysis links in the SNP calendar lead to further details.
- R1b1a1b1a1a2c1a6 FGC5494 2480 BCE
- R1b1a1b1a1a2c1a6. FGC5561 2460 BCE
- R1b1a1b1a1a2c1a6.. FGC19916 1870 BCE
- R1b1a1b1a1a2c1a6... M954/A19914 1660 CE L Cluster 2802 further analysis
- R1b1a1b1a1a2c1a6.... FT63968 1810 CE
- R1b1a1b1a1a2c1a6..... L1314 1910 CE
- R1b1a1b1a1a2c1a6.... FT63968 1810 CE
- R1b1a1b1a1a2c1a6... M954/A19914 1660 CE L Cluster 2802 further analysis
- R1b1a1b1a1a2c1a6.. FGC82708 2430 BCE
- R1b1a1b1a1a2c1a6... FGC69926 2290 BCE
- R1b1a1b1a1a2c1a6.... BY7807 2000 BCE
- R1b1a1b1a1a2c1a6..... BY7804 1680 BCE
- R1b1a1b1a1a2c1a6...... BY11581 1580 BCE
- R1b1a1b1a1a2c1a6....... BY11585 1140 BCE
- R1b1a1b1a1a2c1a6........ BY11577 130 CE L Cluster 2808 further analysis
- R1b1a1b1a1a2c1a6....... BY11585 1140 BCE
- R1b1a1b1a1a2c1a6...... BY11581 1580 BCE
- R1b1a1b1a1a2c1a6..... BY7804 1680 BCE
- R1b1a1b1a1a2c1a6.... BY7807 2000 BCE
- R1b1a1b1a1a2c1a6... FGC69926 2290 BCE
- R1b1a1b1a1a2c1a6.. FGC5495 2450 BCE
- R1b1a1b1a1a2c1a6... FGC5496 2230 BCE
- R1b1a1b1a1a2c1a6.... FGC5512 2100 BCE
- R1b1a1b1a1a2c1a6..... BY3983 2100 BCE L
- R1b1a1b1a1a2c1a6...... A7505 1790 CE L Cluster 2804
- R1b1a1b1a1a2c1a6..... BY3983 2100 BCE L
- R1b1a1b1a1a2c1a6.... PR1289 1960 BCE
- R1b1a1b1a1a2c1a6..... A194 1780 BCE
- R1b1a1b1a1a2c1a6...... CTS2457 250 CE Cluster 2806
- R1b1a1b1a1a2c1a6....... FTD26445 710 CE
- R1b1a1b1a1a2c1a6........ BY101431 1270 CE
- R1b1a1b1a1a2c1a6......... FTD40299 1490 CE
- R1b1a1b1a1a2c1a6........ BY101431 1270 CE
- R1b1a1b1a1a2c1a6....... FTD26445 710 CE
- R1b1a1b1a1a2c1a6...... CTS2457 250 CE Cluster 2806
- R1b1a1b1a1a2c1a6..... A194 1780 BCE
- R1b1a1b1a1a2c1a6.... FGC5512 2100 BCE
- R1b1a1b1a1a2c1a6... FGC5496 2230 BCE
- R1b1a1b1a1a2c1a6.. FGC19916 1870 BCE
- R1b1a1b1a1a2c1a6. FGC5561 2460 BCE
SNP Time and Geography Map  Back to Top
Back to Top
The locations pinned on the map are partly based on the genealogical data provided by project members and partly based on the location data at Scaled Innovation (Rob Spencer). Time period estimates are color coded based on Scaled Innovation.
Modal Haplotypes of FGC5494 Collinses  Back to Top
Back to Top
Four modal haplotypes were generated from the project for Collinses believed to be R-L1314, R-A7505, R-CTS2457, and R-BY11577. An R-FGC5494 modal haplotype in turn was generated from these modals.
| Table #1 Haplotypes for different R-FGC5494 subclade Collinses | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| relative position | 1 | 2 | 3 | 4 | 5-6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14-15 | 16 | 17 | 18 | 19 | 20 | 21 | 22-25 | 26 | 27 | 28-29 | 30 | 31 | 32 | 33 | 34-35 | 36 | 37 | 38 | 39 | 40-41 | 42 | 43 | 44 | 45 | 46 | 47 | 48 | 49-50 | 51 | 52 | 53 | 54 | 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | 63 | 64 | 65 | 66 | 67 | 68 | 69 | 70 | 71 | 72 | 73 | 74 | 75 | 76 | 77 | 78 | 79 | 80 | 81 | 82 | 83 | 84 | 85 | 86 | 87 | 88 | 89 | 90 | 91 | 92 | 93 | 94 | 95 | 96 | 97 | 98 | 99 | 100 | 101 | 102 | 103 | 104 | 105 | 106 | 107 | 108 | 109 | 110 | 111 | relative position | |||||||||
| Short Tandem Repeat Markers (STRS) | DYS393 | DYS390 | DYS19 | DYS391 | DYS385 | DYS426 | DYS388 | DYS439 | DYS389I | DYS392 | DYS389II | DYS458 | DYS459 | DYS455 | DYS454 | DYS447 | DYS437 | DYS448 | DYS449 | DYS464 | DYS460 | Y-GATA-H4 | YCAII | DYS456 | DYS607 | DYS576 | DYS570 | CDY | DYS442 | DYS438 | DYS531 | DYS578 | DYF395S1 | DYS590 | DYS537 | DYS641 | DYS472 | DYF406S1 | DYS511 | DYS425 | DYS413 | DYS557 | DYS594 | DYS436 | DYS490 | DYS534 | DYS450 | DYS444 | DYS481 | DYS520 | DYS446 | DYS617 | DYS568 | DYS487 | DYS572 | DYS640 | DYS492 | DYS565 | DYS710 | DYS485 | DYS632 | DYS495 | DYS540 | DYS714 | DYS716 | DYS717 | DYS505 | DYS556 | DYS549 | DYS589 | DYS522 | DYS494 | DYS533 | DYS636 | DYS575 | DYS638 | DYS462 | DYS452 | DYS445 | Y-GATA-A10 | DYS463 | DYS441 | Y-GGAAT-1B07 | DYS525 | DYS712 | DYS593 | DYS650 | DYS532 | DYS715 | DYS504 | DYS513 | DYS561 | DYS552 | DYS726 | DYS635 | DYS587 | DYS643 | DYS497 | DYS510 | DYS434 | DYS461 | DYS435 | Short Tandem Repeat Markers (STRS) | |||||||||
| member # |
|
member # | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MODAL | 13 | 24 | 14 | 11 | 11-14 | 12 | 12 | 12 | 13 | 13 | 29 | 17 | 9-10 | 11 | 11 | 25 | 15 | 19 | 30 | 15-15-17-17 | 11 | 11 | 19-23 | 16 | 15 | 18 | 17 | 37-39 | 12 | 12 | 11 | 9 | 15-16 | 8 | 10 | 10 | 8 | 10 | 10 | 12 | 23-23 | 17 | 10 | 12 | 12 | 15 | 8 | 12 | 22 | 20 | 13 | 12 | 11 | 13 | 11 | 11 | 12 | 12 | 34 | 15 | 9 | 16 | 12 | 23 | 26 | 19 | 12 | 11 | 13 | 12 | 11 | 9 | 13 | 12 | 10 | 11 | 11 | 30 | 12 | 13 | 24 | 13 | 10 | 10 | 20 | 15 | 17 | 12 | 24 | 17 | 12 | 15 | 24 | 12 | 23 | 18 | 10 | 14 | 17 | 9 | 12 | 11 | MODAL | |||||||||
| L1314-MDL | 14 | 24 | 14 | 11 | 11-14 | 12 | 12 | 12 | 13 | 13 | 29 | 16 | 9-9 | 11 | 11 | 25 | 15 | 19 | 30 | 15-15-15-18 | 11 | 10 | 19-23 | 16 | 15 | 20 | 17 | 37-39 | 12 | 12 | 13 | 9 | 15-16 | 8 | 10 | 10 | 8 | 10 | 10 | 12 | 23-23 | 16 | 10 | 12 | 12 | 15 | 8 | 13 | 22 | 20 | 13 | 12 | 11 | 13 | 10 | 11 | 12 | 12 | 36 | 15 | 9 | 16 | 12 | 23 | 26 | 19 | 11 | 12 | 12 | 11 | 11 | 9 | 13 | 12 | 10 | 11 | 11 | 30 | 12 | 14 | 24 | 13 | 10 | 10 | 21 | 15 | 17 | 13 | 24 | 17 | 13 | 15 | 24 | 12 | 24 | 18 | 10 | 14 | 17 | 9 | 12 | 11 | L1314-MDL | |||||||||
| A7505-MDL | 13 | 24 | 14 | 10 | 11-14 | 12 | 12 | 12 | 13 | 13 | 29 | 17 | 9-10 | 11 | 11 | 23 | 15 | 19 | 29 | 15-15-17-17 | 11 | 11 | 19-23 | 15 | 16 | 18 | 18 | 37-39 | 12 | 12 | 11 | 9 | 16-16 | 8 | 11 | 10 | 8 | 10 | 10 | 12 | 23-23 | 17 | 10 | 12 | 12 | 16 | 8 | 12 | 22 | 20 | 13 | 12 | 11 | 13 | 11 | 11 | 12 | 12 | 34 | 15 | 9 | 16 | 12 | 26 | 26 | 19 | 12 | 11 | 13 | 12 | 12 | 9 | 13 | 12 | 10 | 11 | 11 | 30 | 12 | 13 | 24 | 12 | 10 | 11 | 23 | 15 | 17 | 14 | 24 | 17 | 12 | 15 | 24 | 12 | 24 | 18 | 10 | 14 | 18 | 9 | 12 | 11 | A7505-MDL | |||||||||
| CTS2457-MDL | 13 | 24 | 14 | 11 | 11-15 | 12 | 12 | 12 | 13 | 14 | 29 | 19 | 9-10 | 11 | 11 | 25 | 15 | 19 | 30 | 15-15-17-17 | 11 | 11 | 19-23 | 16 | 16 | 17 | 17 | 36-37 | 12 | 12 | 11 | 9 | 15-16 | 8 | 10 | 10 | 8 | 10 | 10 | 12 | 23-23 | 17 | 11 | 12 | 12 | 15 | 8 | 12 | 22 | 22 | 13 | 13 | 11 | 13 | 11 | 11 | 12 | 12 | 34 | 15 | 9 | 16 | 12 | 27 | 26 | 19 | 12 | 11 | 13 | 12 | 11 | 9 | 13 | 12 | 10 | 11 | 11 | 30 | 12 | 13 | 24 | 13 | 10 | 10 | 20 | 15 | 18 | 12 | 24 | 15 | 12 | 16 | 24 | 12 | 23 | 18 | 10 | 14 | 17 | 9 | 13 | 11 | CTS2457-MDL | |||||||||
| BY11577-MDL | 13 | 24 | 14 | 11 | 12-14 | 12 | 12 | 11 | 12 | 13 | 27 | 17 | 9-10 | 11 | 11 | 25 | 15 | 19 | 30 | 15-15-17-17 | 11 | 11 | 19-23 | 16 | 15 | 18 | 17 | 36-39 | 13 | 12 | 11 | 9 | 15-16 | 8 | 10 | 10 | 8 | 10 | 10 | 12 | 23-23 | 17 | 10 | 12 | 12 | 15 | 8 | 14 | 22 | 20 | 13 | 12 | 11 | 13 | 11 | 11 | 12 | 12 | 34 | 15 | 9 | 16 | 12 | 24 | 26 | 19 | 12 | 11 | 14 | 12 | 11 | 9 | 13 | 12 | 10 | 11 | 11 | 32 | 13 | 13 | 24 | 13 | 10 | 10 | 20 | 15 | 19 | 12 | 24 | 20 | 12 | 16 | 24 | 12 | 23 | 18 | 10 | 14 | 17 | 9 | 12 | 11 | BY11577-MDL | |||||||||
| member # |
| member # | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genetic Distances of FGC5494 Collins Modals 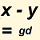 Back to Top
Back to Top
See Genetic Distance in Page Help for details. Note the remarks on the reliability of TMRCA estimation based on STR markers gradually deteriorating as we go further back in time. This would be particularly applicable to modal comparison, as opposed to comparison of individuals.
|
Genetic Distance
Color Key
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
TMRCA Calculations of FGC5494 Collins Modals  Back to Top
Back to Top
See Time to Most Common Recent Ancestor in Page Help for details. Note the remarks on the reliability of TMRCA estimation based on STR markers gradually deteriorating as we go further back in time. This would be particularly applicable to modal comparison, as opposed to comparison of individuals.
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Collins at FTDNA | DNA Portal | Pedigrees | |
| See ABOUT for website information. | |||
| Copyright © 2015 - 2025 collins.dnagen.org. All rights reserved. | |||